This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison.
Future Directions: Elucidating the ABCA1 Membrane Localization Process
Disease-causing ABCA1 mutations occur preferentially in sites that undergo post-translational modifications
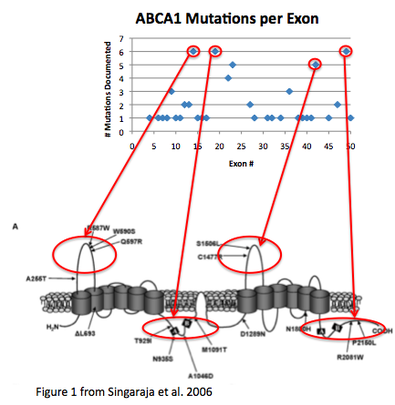
Using data from Singaraja et al. 2006 [1], I looked for a pattern in the distribution of HDL-reducing ABCA1 mutations by plotting mutations against exon number.
I found that exons in the extracellular domains of the protein had the highest number of HDL-reducing mutations.
Not surprisingly, these are the domains that undergo post-translational modifications or bind other molecules (ATP and ApoA-1).
These results agree with the information ABCA1 about that I have found thus far, which suggests post-translational modifications play a major role in the proper localization of ABCA1 to the membrane.
Thus, I chose to look for more clues about how ABCA1 might reach the membrane, starting with other proteins it may interact with on the way there.
I found that exons in the extracellular domains of the protein had the highest number of HDL-reducing mutations.
Not surprisingly, these are the domains that undergo post-translational modifications or bind other molecules (ATP and ApoA-1).
These results agree with the information ABCA1 about that I have found thus far, which suggests post-translational modifications play a major role in the proper localization of ABCA1 to the membrane.
Thus, I chose to look for more clues about how ABCA1 might reach the membrane, starting with other proteins it may interact with on the way there.
CD36 appears to influence ABCA1 membrane localization
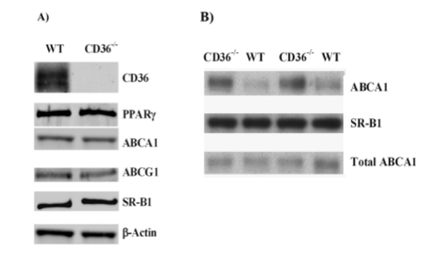
Figure 6 a, b from [2]
CD36 is a transmembrane platelet glycoprotein that is believed to interact with the golgi body. It is in the same protein family as the HDL receptor protein SR-B1.
In mice that lacked the CD36 gene (CD36-/-), ABCA1 membrane localization (panel B), but not protein levels (panel B), increased compared to normal (WT) mouse cells. Darker bands in the Western blots shown at right indicate higher amounts of protein [2].
This indicates that CD36 or its targets may play an uncharacterized role in the localization of ABCA1 to the plasma membrane. More specifically, it may inhibit ABCA1 localization somehow.
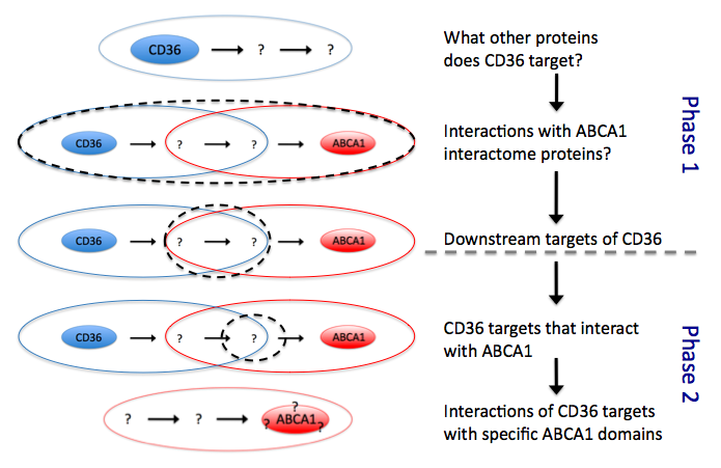
Experiment overview
To further explore any possible connections between CD36 and ABCA1, I suggest attempting to identify any proteins that act downstream of CD36, but upstream of ABCA1.
HYPOTHESIS: Downstream targets of CD36 negatively regulate ABCA1 localization
Experiment 1a: Tandem affinity purification
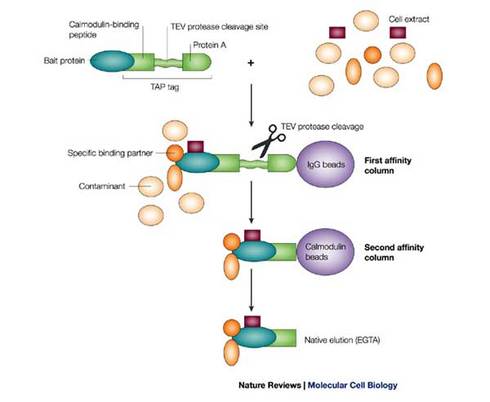
Tandem affinity purification schematic [3]
PURPOSE: Isolate proteins that form complexes with CD36 to expand its interactome
Tandem affinity purification is a method for isolating and identifying proteins in complexes.
In brief, this technique uses a series of affinity assays to first pull out binding proteins from a cellular extract, and then in several chemical steps, remove contaminants and groups used in the isolation process so that the proteins of interest can be sequenced by mass spectrometry [3].
Tandem affinity purification is a method for isolating and identifying proteins in complexes.
In brief, this technique uses a series of affinity assays to first pull out binding proteins from a cellular extract, and then in several chemical steps, remove contaminants and groups used in the isolation process so that the proteins of interest can be sequenced by mass spectrometry [3].
Experiment 1b: Protein microarrays

An image of a microarray chip [4]
PURPOSE: Identify other proteins affected by varying the levels of CD36 (downstream of CD36 in pathway)
Microarrays work by chemically attaching a DNA, RNA or protein probe (a short molecule) to a small area on a chip. The chip is then exposed to cellular extracts, and molecules in the extract are captured by the appropriate probe. Finally, the amount of molecules bound to the probes can be determined based on the intensity of the fluorescent signal given off by each spot on the array after the target proteins bind [5].
A protein microarray can be used to compare the expression levels of hundreds of proteins under different biological conditions. This is very helpful for clarifying functional relationships between proteins.
In the case of this experiment, I would compare expression of all known ABCA1 and CD36 interaction partners (as identified by experiment 1a) in macrophages that were:
1. Homozygous for a CD36 null allele
2. Heterozygous for the null allele
3. Homozygous for a normal CD36 allele
Microarrays work by chemically attaching a DNA, RNA or protein probe (a short molecule) to a small area on a chip. The chip is then exposed to cellular extracts, and molecules in the extract are captured by the appropriate probe. Finally, the amount of molecules bound to the probes can be determined based on the intensity of the fluorescent signal given off by each spot on the array after the target proteins bind [5].
A protein microarray can be used to compare the expression levels of hundreds of proteins under different biological conditions. This is very helpful for clarifying functional relationships between proteins.
In the case of this experiment, I would compare expression of all known ABCA1 and CD36 interaction partners (as identified by experiment 1a) in macrophages that were:
1. Homozygous for a CD36 null allele
2. Heterozygous for the null allele
3. Homozygous for a normal CD36 allele
Experiment 2a: Yeast two-hybrid screen #1
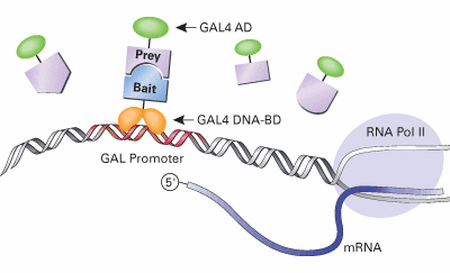
Y2H schematic [5]
PURPOSE: Test for interactions between CD36 targets and known ABCA1 interaction partners
Yeast two-hybrid screens test for protein-protein interactions by engineering "bait" and "prey" chimeric proteins expressed in yeast. The yeast will only be able to perform transcription (e.g. of an enzyme) when the "bait" protein bound to the promoter binds to a "prey" protein that binds DNA at another location to allow formation of a transcription complex.
The yeast are exposed to libraries of "prey" proteins. Interactions between the "bait" and "prey" are indicated by transcription of the genes that require the promoter in question [5].
Experiment 2b: Yeast two-hybrid screen #2
PURPOSE: Test for interactions between proteins intermediate to CD36 and ABCA1 and different alleles of ABCA1
This step will help identify which ABCA1 domains interact with the proteins affected by CD36 levels and may be involved in localization to the plasma membrane.
This step will help identify which ABCA1 domains interact with the proteins affected by CD36 levels and may be involved in localization to the plasma membrane.
Next steps
If this experiment succeeded in identifying domains of ABCA1 that interact with downstream targets of CD36, I would propose investigating the in vivo consequences of knocking out these proteins using RNA interference by...
1. Looking for the effects on other proteins in the ABCA1 interactome using microarrays
2. Quantifying ABCA1 plasma membrane localization in macrophages
Finally, if knockout mice could be generated for these proteins, I would also want to examine the effects of knocking out the proteins of interest on plasma HDL levels in homozygous mutant mice.
1. Looking for the effects on other proteins in the ABCA1 interactome using microarrays
2. Quantifying ABCA1 plasma membrane localization in macrophages
Finally, if knockout mice could be generated for these proteins, I would also want to examine the effects of knocking out the proteins of interest on plasma HDL levels in homozygous mutant mice.
Sources:
1. Singaraja R, Visscher H, James ER, Chroni A, Coutinho JM, Brunham LR, Kang MH, Zannis VI, Chimini G and Hayden MR. Lipid Phenotypes Both In Vivo and In Vitro Specific Mutations in ABCA1 Have Discrete Effects on ABCA1 Function and Lipid Phenotypes Both In Vivo and In Vitro. Circ. Res. 2006; 99: 389-397.
2. Yue P, Chen Z, Nassir F, Bernal-Mizrachi C, Finck B, Azhar S, and Abumrad N. Enhanced Hepatic apoA-I Secretion and Peripheral Efflux of Cholesterol and Phospholipid in CD36 Null Mice. PLoS One. 2010; 5(3): e9906.
3. Huber LA. Is proteomics heading in the wrong direction? Nature Reviews Molecular Cell Biology. 2003; 4: 74-80
4. http://publish.uwo.ca/~bneff/research_eefg.htm
5. Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999 Jan;21(1 Suppl):33-7.
1. Singaraja R, Visscher H, James ER, Chroni A, Coutinho JM, Brunham LR, Kang MH, Zannis VI, Chimini G and Hayden MR. Lipid Phenotypes Both In Vivo and In Vitro Specific Mutations in ABCA1 Have Discrete Effects on ABCA1 Function and Lipid Phenotypes Both In Vivo and In Vitro. Circ. Res. 2006; 99: 389-397.
2. Yue P, Chen Z, Nassir F, Bernal-Mizrachi C, Finck B, Azhar S, and Abumrad N. Enhanced Hepatic apoA-I Secretion and Peripheral Efflux of Cholesterol and Phospholipid in CD36 Null Mice. PLoS One. 2010; 5(3): e9906.
3. Huber LA. Is proteomics heading in the wrong direction? Nature Reviews Molecular Cell Biology. 2003; 4: 74-80
4. http://publish.uwo.ca/~bneff/research_eefg.htm
5. Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999 Jan;21(1 Suppl):33-7.