This web page was produced as an assignment for Genetics 677, an undergraduate course at UW‐Madison.
Microarray data
About microarrays
Microarrays can be used to quantify the expression of mRNA transcripts (the intermediates between genes and their protein products) under different conditions. For example, one common application is to compare mRNA expression in a disease patient to that of a healthy person.
This is accomplished by binding DNA oligonucleotides that are complementary to the mRNA of interest, and allowing mRNA from various samples to hybridize to these "probes". After hybridization, the quantity of bound mRNA can be ascertained from the strength of a fluorescent signal at each probe that is proportional to the amount of hybridized mRNA.
This is accomplished by binding DNA oligonucleotides that are complementary to the mRNA of interest, and allowing mRNA from various samples to hybridize to these "probes". After hybridization, the quantity of bound mRNA can be ascertained from the strength of a fluorescent signal at each probe that is proportional to the amount of hybridized mRNA.
ABCA1 expression in a Tangier disease patient quantified by cDNA microarray
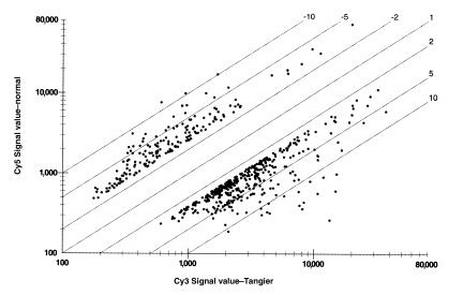
mRNA levels in a Tangier disease patient (horizontal axis) and normal cells (vertical axis) [1]
Microarrays were used to help identify ABCA1 as the disease locus for Tangier disease [1].
The chart at left shows abnormally high and low expression of approximately 500 genes in a TD patient's cells compared to normal cells.
The chart at left shows abnormally high and low expression of approximately 500 genes in a TD patient's cells compared to normal cells.
NCBI GEO Profiles database

http://www.ncbi.nlm.nih.gov/sites/geo [2]
NCBI's Gene Expression Omnibus is a database of gene expression data.
I was unable to find microarray data on GEO for human ABCA1 expression in Tangier disease patients. However, there are data for ABCA1 expression in several other human diseases and Mus musculus disease models. There are also data for expression of many other genes in familial hypercholesterolemia, which can be accessed here.
I was unable to find microarray data on GEO for human ABCA1 expression in Tangier disease patients. However, there are data for ABCA1 expression in several other human diseases and Mus musculus disease models. There are also data for expression of many other genes in familial hypercholesterolemia, which can be accessed here.
Sources:
1. Rust S. et al. Tangier disease is caused by mutations in the gene encoding ATP‐binding cassette transporter 1. Nat Genet. 1999 Aug; 22(4):316–8.
2. http://www.ncbi.nlm.nih.gov/sites/geo
1. Rust S. et al. Tangier disease is caused by mutations in the gene encoding ATP‐binding cassette transporter 1. Nat Genet. 1999 Aug; 22(4):316–8.
2. http://www.ncbi.nlm.nih.gov/sites/geo
